|
T. Wiedmer, S.T. Teoh, E. Christodoulaki, G. Wolf, C. Tian, V. Sedlyarov, A. Jarret, P. Leippe, F. Frommelt, A. Ingles-Prieto, S. Lindinger, B.M.G. Barbosa, S. Onstein, C. Klimek, J. Garcia, I. Serrano, D. Reil, D. Santacruz, M. Piotrowski, S. Noell, C. Bueschl, H. Li, G. Chi, S. Mereiter, T. Oliveira, J.M. Penninger, D.B. Sauer, C.M. Steppan, C. Viollet, K. Klavins, J.T. Hannich, U. Goldmann, G. Superti-Furga. Metabolic mapping of the human solute carrier superfamily. Mol Syst Biol. 21: 560-598 (2025). [Link] |
|
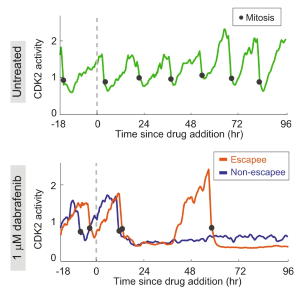
T.E. Hoffman,* C. Tian,* V. Nangia,* C. Yang, S. Regot, L. Gerosa, S.L. Spencer. CDK2 activity crosstalk on the ERK kinase translocation reporter can be resolved computationally. Cell Syst. 16: 101162 (2025). [Link] |
|
K.R. Jacobson, A.M. Saleh, S.N. Lipp, C. Tian, A.R. Watson, C.M. Luetkemeyer, A.R. Ocken, S.L. Spencer, T.L. Kinzer-Ursem, S. Calve. Extracellular matrix protein composition dynamically changes during murine forelimb development. iScience. 27: 108838 (2024). [Link] |
|
J.Y. Chen, C. Hug, J. Reyes, C. Tian, L. Gerosa, F. Fröhlich, B. Ponsioen, H.J.G. Snippert, S.L. Spencer, A. Jambhekar, P.K. Sorger, G. Lahav. Multi range ERK responses shape the proliferative trajectory of single cells following oncogene induced senescence. Cell. Rep. 42: 112252 (2023). [Link] |
| C. Yang,* C. Tian,* T. Hoffman,* N. Jacobsen, S. Spencer. Melanoma subpopulations that rapidly escape MAPK pathway inhibition incur DNA damage and rely on stress signalling. Nat. Commun. 12: 1747 (2021). (*: equal contribution) [Link] | |
| C. Tian,* C. Yang,* S. Spencer. EllipTrack: a global-local cell-tracking pipeline for 2D fluorescence time-lapse microscopy. Cell. Rep. 32: 107984 (2020). [Link] | |
|
M. Min, Y. Rong, C. Tian, S. Spencer. Temporal integration of mitogen history in mother cells controls proliferation of daughter cells. Science. 368: aay8241 (2020). [Link] |
|
R. Fu, A. Gillen, R. Sheridan, C. Tian, M. Daya, Y. Hao, J. Hesselberth, K. Riemondy. clustifyr: An R package for automated single-cell RNA sequencing cluster classification. F1000Res. 9: 223 (2020). [Link] |
|
I. Miller, M. Min, C. Yang, C. Tian, S. Gookin, D. Carter, S. Spencer. Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell. Rep. 24: 1105-1112 (2018). [Link] |
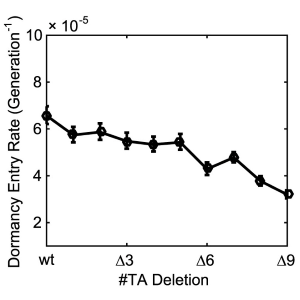
| C. Tian, S. Semsey, N. Mitarai. Synchronized switching of multiple Toxin–Antitoxin modules by (p) ppGpp fluctuation. Nucleic. Acids. Res. 45: 8180-8189 (2017). [Link] | |
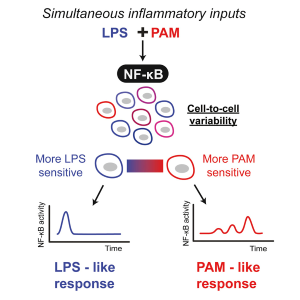
| R.A. Kellogg,* C. Tian,* M. Etzrodt, S. Tay. Cellular Decision Making by Non-Integrative Processing of TLR Inputs. Cell. Rep. 19: 125-135 (2017). [Link] | |
| C. Tian,* M. Roghanian,* M.G. Jorgensen, K. Sneppen, M.A. Sørensen, K. Gerdes, N. Mitarai. Rapid curtailing of the stringent response by Toxin-Antitoxin-encoded mRNases. J. Bacteriol. 198: 1918-1926 (2016). [Link] | |
| C. Tian, N. Mitarai. Bifurcation of transition paths induced by coupled bistable systems. J. Chem. Phys. 144: 215102 (2016). [Link] | |
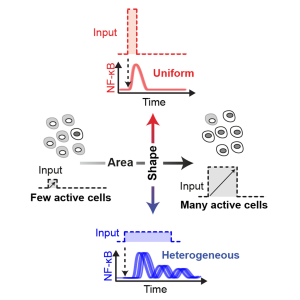
| R.A. Kellogg, C. Tian, T. Lipniacki, S.R. Quake, S. Tay. Digital signaling decouples activation probability and population heterogeneity. eLife 4: e08931 (2015). [Link] | |
|
H. Zhang, M. Lin, H. Shi, W. Ji, L. Huang, X. Zhang, S. Shen, R. Gao, S. Wu, C. Tian, Z. Yang, G. Zhang, S. He, H. Wang, T. Saw, Y. Chen, Q. Ouyang. Programming a Pavlovian-like conditioning circuit in Escherichia coli. Nat. Commun. 5:3102 (2014). [Link] |